Team members
Faculty
Mani Srivastava (Thrust 3 Lead, UCLA )
Timothy Hnat (Chief Software Architect, Memphis)
Tyson Condie (UCLA)
Simone Carini (UCSF)
Santosh Kumar (Memphis)
Syed Monowar Hossain (Memphis)
Nasir Ali (Memphis)
Nusrat Nasrin (Memphis).
Students, Post Docs
Bo-Jhang Ho (UCLA)
Matteo Interlandi (UCLA)
Addison Mayberry (UMass)
Mobile Sensor Big Data Architecture
Development and validation of any new mHealth biomarker requires conducting research studies in lab and field settings to collect raw sensor data with appropriate labels (e.g., self-reports). A general-purpose software platform that can enable such data collection consists of software on sensors, mobile phones, and the cloud, which all need to work together. Each of these software must be modular so as to enable seamless mix-and=match to customize it for various study needs. The software architecture for such a platform needs several attributes.
First, it must support concurrent connections to a wide variety of high-rate wearable sensors with an ability to plug-in new sensors.
Second, all three platforms must ingest the large volume of rapidly arriving data for which native support does not yet exist in the smartphone hardware or operating system without falling behind and losing data.
Third, it needs to support reliable storage of a quickly-growing volume of sensor data, the archival of which is critical to the development and validation of new biomarkers.
Fourth, it is desirable to quickly analyze incoming data to monitor signal quality so that any errors in sensor attachment or placement can be promptly fixed to maximize data yield.
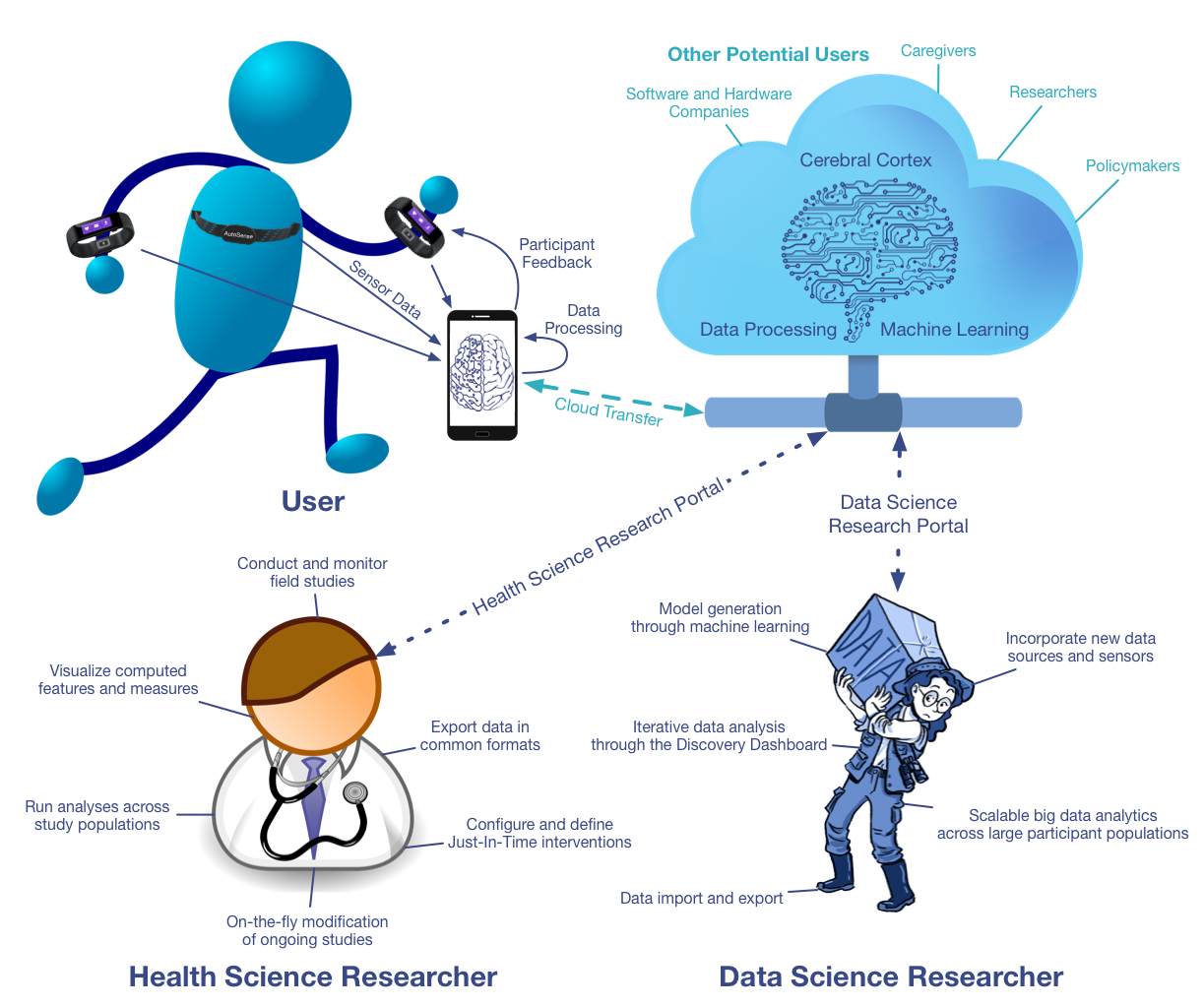
System Overview
There are three core users of mCerebrum and Cerebral Cortex. (1) The user wearing sensors and interacting with mCerebrum which uploads data to Cerebral Cortex, (2) the health science researcher that conduct studies, visualize field data, run population-scale analysis, and (3) the data science researcher that constructs models through machine learning, runs interative analysis through web-based dashboards, and buils scalable data analytics across large populations. (Click on image for larger version)
Fifth, the smartphone and/or the cloud needs to support the sense-analyze-act pipeline for high-rate streaming sensor data. This is necessary to prompt self-reports (for collection of labels) as well as confirm/refute prompts for validation of new biomarkers in the field. Sense-analyze-act support is also needed to aid development and evaluation of sensor-triggered interventions.
Sixth, it needs seamless sharing of streaming data from multiple sensors to enable computation of multi-sensor biomarkers (e.g., stress, smoking, eating).
Seventh, the platform needs to be general-purpose and extensible to support a wide variety of sensors, biomarkers, and study designs.
Eighth, it needs to be architecturally scalable so that it can support concurrent computation of a large number of biomarkers (each of which requires complex processing) without saturating the computational capacity or depleting the battery life of the smartphone.
Ninth, the smartphone platform needs to carefully control interruptions to study participants from various sources (e.g., self-report, ecological momentary assessment (EMA) and interventions (EMI), fixing sensor attachments) limiting user burden and cognitive overload while satisfying the numerous study requirements.
Tenth, the cloud platform must support concurrent data collection from hundreds, if not thousands of smartphone instances deployed in the field and reliably offload raw sensor data, derived features and biomarkers, and self-reports.
Eleventh, the cloud platform needs to provide a dashboard to remotely monitor the quality of data collection and participant compliance so as to intervene when necessary to ensure high data-yield.
Twelfth, for mobile sensor big data analytics, the cloud platform must support export of sensor data, features, biomarkers, and self-reports for population-scale analysis, as well as offer exploratory visualization and analysis. Last, but not least, the cloud platform must support annotation of data with metadata and provenance information so as to enable comparative analysis, reproducibility, and third party research.
The big data computing architectures for all three platforms (sensors, smartphones, and cloud) by MD2K are aimed at meeting all of the above requirements. The publications listed below provide details of these architectures.
Publications
Bo-Jhang Ho, Bharathan Balaji, Nima Nikzad and Mani Srivastava.
Emu: Engagement Modeling for User Studies. In UbiTention 2017: 2nd International Workshop on Smart & Ambient Notification and Attention Management. 2017. URL BibTeX@inproceedings{ho2017emu, author = "Bo-Jhang Ho and Bharathan Balaji and Nima Nikzad and Mani Srivastava", title = "Emu: Engagement Modeling for User Studies", booktitle = "UbiTention 2017: 2nd International Workshop on Smart \& Ambient Notification and Attention Management", year = 2017, organization = "ACM", abstract = "Mobile technologies that drive just-in-time ecological momentary assessments and interventions provide an unprecedented view into user behaviors and opportunities to manage chronic conditions. The success of these methods rely on engaging the user at the appropriate moment, so as to maximize questionnaire and task completion rates. However, mobile operating systems provide little support to precisely specify the contextual conditions in which to notify and engage the user, and study designers often lack the expertise to build context-aware software themselves. To address this problem, we have developed Emu, a framework that eases the development of context-aware study applications by providing a concise and powerful interface for specifying temporal- and contextual-constraints for task notifications. In this paper we present the design of the Emu API and demonstrate its use in capturing a range of scenarios common to smartphone-based study applications.", pubstate = "published", tppubtype = "inproceedings", url = "https://md2k.org/images/papers/software/UbiTtention2-13.pdf" }Syed Monowar Hossain, Timothy Hnat, Nazir Saleheen, Nusrat Jahan Nasrin, Joseph Noor, Bo-Jhang Ho, Tyson Condie, Mani Srivastava and Santosh Kumar.
mCerebrum: An mHealth Software Platform for Development and Validation of Digital Biomarkers and Interventions. In The ACM Conference on Embedded Networked Sensor Systems (SenSys). 2017. URL BibTeX@inproceedings{hossain2017mcerebrum, author = "Syed Monowar Hossain and Timothy Hnat and Nazir Saleheen and Nusrat Jahan Nasrin and Joseph Noor and Bo-Jhang Ho and Tyson Condie and Mani Srivastava and Santosh Kumar", title = "mCerebrum: An mHealth Software Platform for Development and Validation of Digital Biomarkers and Interventions", booktitle = "The ACM Conference on Embedded Networked Sensor Systems (SenSys)", year = 2017, month = "", organization = "ACM", abstract = "The development and validation studies of new multisensory biomark-ers and sensor-triggered interventions requires collecting raw sen-sor data with associated labels in the natural field environment. Unlike platforms for traditional mHealth apps, a software platform for such studies needs to not only support high-rate data ingestion, but also share raw high-rate sensor data with researchers, while supporting high-rate sense-analyze-act functionality in real-time. We present mCerebrum, a realization of such a platform, which supports high-rate data collections from multiple sensors with real-time assessment of data quality. A scalable storage architecture (with near optimal performance) ensures quick response despite rapidly growing data volume. Micro-batching and efficient sharing of data among multiple source and sink apps allows reuse of com-putations to enable real-time computation of multiple biomarkers without saturating the CPU or memory. Finally, it has a reconfig-urable scheduler which manages all prompts to participants that is burden- and context-aware. With a modular design currently span-ning 23+ apps, mCerebrum provides a comprehensive ecosystem of system services and utility apps. The design of mCerebrum has evolved during its concurrent use in scientific field studies at ten sites spanning 106,806 person days. Evaluations show that com-pared with other platforms, mCerebrum’s architecture and design choices support 1.5 times higher data rates and 4.3 times higher storage throughput, while causing 8.4 times lower CPU usage.", pubstate = "published", tppubtype = "inproceedings", url = "https://md2k.org/images/papers/software/mCerebrum-SenSys-2017.pdf" }Timothy Hnat, Syed Hossain, Nasir Ali, Simona Carini, Tyson Condie, Ida Sim, Mani Srivastava and Santosh Kumar.
mCerebrum and Cerebral Cortex: A Real-time Collection, Analytic, and Intervention Platform for High-frequency Mobile Sensor Data. In AMIA (American Medical Informatics Association) 2017 Annual Symposium. 2017. BibTeX@inproceedings{hnat2017mcerebrum, author = "Timothy Hnat and Syed Hossain and Nasir Ali and Simona Carini and Tyson Condie and Ida Sim and Mani Srivastava and Santosh Kumar", title = "mCerebrum and Cerebral Cortex: A Real-time Collection, Analytic, and Intervention Platform for High-frequency Mobile Sensor Data", booktitle = "AMIA (American Medical Informatics Association) 2017 Annual Symposium", year = 2017, organization = "American Medical Informatics Association", abstract = "The Center of Excellence for Mobile Sensor Data to Knowledge (MD2K)1 has developed two open-source software platforms2 (for mobile phones and the cloud) that enables the collection of high-frequency raw sensor data (at 800+ Hz for 70+ million samples/day), curation, analytics, storage ( 2GB/day) and secure uploads to a cloud. mCere-brum supports concurrent collection of streaming data from multiple devices (including wearables: Microsoft Band, research-grade MotionSenseHRV, EasySense, and AutoSense), phone sensors (e.g., GPS), Omron scale and blood pressure monitors and a smart toothbrush. mCerebrum continuously assesses data quality to detect issues of sensor detachment or placement on the body so they can be addressed. Data science research conducted by MD2K has resulted in 10 mHealth biomarkers: stress likelihood, smoking via hand gestures, nicotine craving, eating, lung con-gestion, heart motion, location, physical activity, driving, and drug use. Several of these biomarkers are computed in real-time on the phone to support biomarker-triggered Just-in-Time Adaptive Interventions (JITAI). Cerebral Cortex is the big data companion of mCerebrum designed to support population-scale data analysis, visu-alization, and model development. It currently supports thousands of concurrent mCerebrum instances and provides machine-learning model development capabilities on population-scale data sets. Cerebral Cortex supports the same mHealth biomarkers that our phone platform computes, but does so on the entire dataset using an Apache Spark pow-ered abstraction that is designed for batch-mode computations as well as near real-time analysis via a data science dashboard. The MD2K platform uses Open mHealth mobile data exchange standards that are being harmonized with FHIR for EHR data exchange. All MD2K software is available under the BSD 2-Clause license3. By collecting and storing high-frequency raw sensor data, our approach enables external validation of computed biomarkers as well as computation of new biomarkers in the future. This benefit is akin to how biomedical stud-ies archive biospecimens in biobanks so that they can be reprocessed to take advantage of future improvements in assays and support discoveries not possible at the time of data collection. Our platform addresses the challenges of high frequency, large volume, rapid variability and battery life limitations to enable long-lasting digital biobanks or digibanks as research utilities for mobile-driven translational research. This demonstration will showcase data collection and visualization from multiple wearable sensor platforms and just-in-time user engagement will demonstrate how the platform can react in real-time to biomarker events. The demon-stration will show how Cerebral Cortex’s capabilities for near real-time visualization of mCerebrum data are currently utilized to manage multiple concurrent clinical study participants at several sites across the United States. Finally, we will explore the interactive data science interface and showcase biomarker generation and model evaluation.", pubstate = "published", tppubtype = "inproceedings" }Barbara E Bierer, Rebecca Li, Mark Barnes and Ida Sim.
A Global, Neutral Platform for Sharing Trial Data. New England Journal of Medicine, 2016. URL BibTeX@article{Bierer2016, author = "Barbara E Bierer and Rebecca Li and Mark Barnes and Ida Sim", title = "A Global, Neutral Platform for Sharing Trial Data", journal = "New England Journal of Medicine", year = 2016, abstract = "Sharing clinical trial data is critical in order to inform clinical and regulatory decision making and honor trial partici-pants who put themselves at risk to advance science. A recent In-stitute of Medicine (IOM) report argues that availability of de-identified (anonymized) patient-level data from clinical trials can permit verification of original results, enhancing public trust and accountability; facilitate other critical research (e.g., evaluation of adverse event rates according to compound class or subpopula-tion or identification of surro-gate end points); and avert dupli-cate trials, shielding participants from unnecessary risk.1 If such goals are to be achieved, patient-level data must be readily find-able and available for aggrega-tion and analysis across multiple sources to enable the widest range of secondary research uses.", publisher = "Massachusetts Medical Society", pubstate = "published", tppubtype = "article", url = "https://md2k.org/images/papers/software/nejmp1605348_Sim.pdf" }Muhammad Ali Gulzari, Matteo Interlandi, Seunghyun Yoo, Sai Deep Tetali, Tyson Condie, Todd Millstein and Miryung Kim.
BigDebug: Debugging Primitives for Interactive Big Data Processing in Spark. In Proceedings of the 38th International Conference on Software Engineering. 2016, 784–795. URL, DOI BibTeX@inproceedings{Gulzar:2016:BDP:2884781.2884813, author = "Muhammad Ali Gulzari and Matteo Interlandi and Seunghyun Yoo and Sai Deep Tetali and Tyson Condie and Todd Millstein and Miryung Kim", title = "BigDebug: Debugging Primitives for Interactive Big Data Processing in Spark", booktitle = "Proceedings of the 38th International Conference on Software Engineering", year = 2016, series = "ICSE '16", pages = "784--795", address = "Austin, Texas", publisher = "ACM", abstract = "Developers use cloud computing platforms to process a large quantity of data in parallel when developing big data analytics. Debugging the massive parallel computations that run in today's datacenters is time consuming and error-prone. To address this challenge, we design a set of interactive, real-time debugging primitives for big data processing in Apache Spark, the next generation data-intensive scalable cloud computing platform. This requires rethinking the notion of step-through debugging in a traditional debugger such as gdb, because pausing the entire computation across distributed worker nodes causes significant delay and naively inspecting millions of records using a watchpoint is too time consuming for an end user. First, BigDebug's simulated breakpoints and on-demand watchpoints allow users to selectively examine distributed, intermediate data on the cloud with little overhead. Second, a user can also pinpoint a crash-inducing record and selectively resume relevant sub-computations after a quick fix. Third, a user can determine the root causes of errors (or delays) at the level of individual records through a fine-grained data provenance capability. Our evaluation shows that BigDebug scales to terabytes and its record-level tracing incurs less than 25% overhead on average. It determines crash culprits orders of magnitude more accurately and provides up to 100% time saving compared to the baseline replay debugger. The results show that BigDebug supports debugging at interactive speeds with minimal performance impact.", doi = "10.1145/2884781.2884813", isbn = "978-1-4503-3900-1", keywords = "big data analytics, data-intensive scalable computing (DISC), Debugging, fault localization and recovery, interactive tools", pubstate = "published", tppubtype = "inproceedings", url = "https://md2k.org/images/papers/software/p784-gulzar_bigdebug16.pdf" }Cheng Zhang, Junrui Yang, Caleb Southern, Thad E Starner and Gregory D Abowd.
WatchOut: Extending Interactions on a Smartwatch with Inertial Sensing. In Proceedings of the 2016 ACM International Symposium on Wearable Computers. 2016, 136–143. URL, DOI BibTeX@inproceedings{Zhang:2016:WEI:2971763.2971775, author = "Zhang, Cheng and Yang, Junrui and Southern, Caleb and Starner, Thad E. and Abowd, Gregory D.", title = "WatchOut: Extending Interactions on a Smartwatch with Inertial Sensing", booktitle = "Proceedings of the 2016 ACM International Symposium on Wearable Computers", year = 2016, series = "ISWC '16", pages = "136--143", address = "New York, NY, USA", publisher = "ACM", abstract = "Current interactions on a smartwatch are generally limited to a tiny touchscreen, physical buttons or knobs, and speech. We present WatchOut, a suite of interaction techniques that includes three families of tap and swipe gestures which extend input modalities to the watch's case, bezel, and band. We describe the implementation of a user-independent gesture recognition pipeline based on data from the watch's embedded inertial sensors. In a study with 12 participants using both a round- and square-screen watch, the average gesture classification accuracies ranged from 88.7% to 99.4%. We demonstrate applications of this richer interaction capability, and discuss the strengths, limitations, and future potential for this work.", acmid = 2971775, doi = "10.1145/2971763.2971775", isbn = "978-1-4503-4460-9", keywords = "inertial sensing, machine learning, mobile interactions, smartwatch", location = "Heidelberg, Germany", numpages = 8, url = "https://md2k.org/images/papers/software/p136-zhang_watchout" }Gabriel Reyes, Dingtian Zhang, Sarthak Ghosh, Pratik Shah, Jason Wu, Aman Parnami, Bailey Bercik, Thad Starner, Gregory D Abowd and Keith W Edwards.
Whoosh: Non-Voice Acoustics for Low-Cost, Hands-Free, and Rapid Input on Smartwatches. In Proceedings of the 2016 ACM International Joint Conference on Pervasive and Ubiquitous Computing (to appear). 2016. URL BibTeX@inproceedings{Reyes2016, author = "Gabriel Reyes and Dingtian Zhang and Sarthak Ghosh and Pratik Shah and Jason Wu and Aman Parnami and Bailey Bercik and Thad Starner and Gregory D. Abowd and W. Keith Edwards", title = "Whoosh: Non-Voice Acoustics for Low-Cost, Hands-Free, and Rapid Input on Smartwatches", booktitle = "Proceedings of the 2016 ACM International Joint Conference on Pervasive and Ubiquitous Computing (to appear)", year = 2016, abstract = "We present an alternate approach to smartwatch interactions using non-voice acoustic input captured by the device’s microphone to complement touch and speech. Whoosh is an interaction technique that recognizes the type and length of acoustic events performed by the user to enable low-cost, hands-free, and rapid input on smartwatches. We build a recognition system capable of detecting non-voice events directed at and around the watch, including blows, sip-and-puff, and directional air swipes, without hardware modifications to the device. Further, inspired by the design of musical instruments, we develop a custom modification of the physical structure of the watch case to passively alter the acoustic response of events around the bezel; this physical redesign expands our input vocabulary with no additional electronics. We evaluate our technique across 8 users with 10 events exhibiting up to 90.5% ten-fold cross validation accuracy on an unmodified watch, and 14 events with 91.3% ten-fold cross validation accuracy with an instrumental watch case. Finally, we share a number of demonstration applications, including multi-device interactions, to highlight our technique with a real-time recognizer running on the watch.", keywords = "hands-free, interaction techniques, Interfaces, non-voice acoustics, on-body input, smartwatches, wearable computing", pubstate = "published", tppubtype = "inproceedings", url = "https://md2k.org/images/papers/software/p120-reyes_whoosh.pdf" }Barbara E Bierer, Rebecca Li, Mark Barnes and Ida Sim.
A Global, Neutral Platform for Sharing Trial Data. New England Journal of Medicine, 2016. URL BibTeX@article{bierer2016global, author = "Barbara E Bierer and Rebecca Li and Mark Barnes and Ida Sim", title = "A Global, Neutral Platform for Sharing Trial Data", journal = "New England Journal of Medicine", year = 2016, abstract = "Sharing clinical trial data is critical in order to inform clinical and regulatory decision making and honor trial partici-pants who put themselves at risk to advance science. A recent In-stitute of Medicine (IOM) report argues that availability of de-identified (anonymized) patient-level data from clinical trials can permit verification of original results, enhancing public trust and accountability; facilitate other critical research (e.g., evaluation of adverse event rates according to compound class or subpopula-tion or identification of surro-gate end points); and avert dupli-cate trials, shielding participants from unnecessary risk.1 If such goals are to be achieved, patient-level data must be readily find-able and available for aggrega-tion and analysis across multiple sources to enable the widest range of secondary research uses.", publisher = "Massachusetts Medical Society", pubstate = "published", tppubtype = "article", url = "https://md2k.org/images/papers/software/nejmp1605348_Sim.pdf" }Markus Weimer, Yingda Chen, Byung-Gon Chun, Tyson Condie, Carlo Curinoo, Chris Douglas, Yunseong Lee, Tony Majestro, Dahlia Malkhi, Sergiy Matusevych, Brandon Myers, Shravan Narayanamurthy, Raghu Ramakrishnan, Sriram Rao, Russel Sears, Beysim Sezgin and Julia Wang.
REEF: Retainable Evaluator Execution Framework. In Proceedings of the 2015 ACM SIGMOD International Conference on Management of Data. 2015, 1343–1355. URL, DOI BibTeX@inproceedings{Weimer:2015:RRE:2723372.2742793, author = "Markus Weimer and Yingda Chen and Byung-Gon Chun and Tyson Condie and Carlo Curinoo and Chris Douglas and Yunseong Lee and Tony Majestro and Dahlia Malkhi and Sergiy Matusevych and Brandon Myers and Shravan Narayanamurthy and Raghu Ramakrishnan and Sriram Rao and Russel Sears and Beysim Sezgin and Julia Wang", title = "REEF: Retainable Evaluator Execution Framework", booktitle = "Proceedings of the 2015 ACM SIGMOD International Conference on Management of Data", year = 2015, series = "SIGMOD '15", pages = "1343--1355", address = "Melbourne, Victoria, Australia", publisher = "ACM", abstract = "In this demo proposal, we describe REEF, a framework that makes it easy to implement scalable, fault-tolerant runtime environments for a range of computational models. We will demonstrate diverse workloads, including extract-transform-load MapReduce jobs, iterative machine learning algorithms, and ad-hoc declarative query processing. At its core, REEF builds atop YARN (Apache Hadoop 2's resource manager) to provide retainable hardware resources with lifetimes that are decoupled from those of computational tasks. This allows us to build persistent (cross-job) caches and cluster-wide services, but, more importantly, supports high-performance iterative graph processing and machine learning algorithms. Unlike existing systems, REEF aims for composability of jobs across computational models, providing significant performance and usability gains, even with legacy code. REEF includes a library of interoperable data management primitives optimized for communication and data movement (which are distinct from storage locality). The library also allows REEF applications to access external services, such as user-facing relational databases. We were careful to decouple lower levels of REEF from the data models and semantics of systems built atop it. The result was two new standalone systems: Tang, a configuration manager and dependency injector, and Wake, a state-of-the-art event-driven programming and data movement framework. Both are language independent, allowing REEF to bridge the JVM and .NET.", doi = "10.1145/2723372.2742793", isbn = "978-1-4503-2758-9", keywords = "Big Data, databases, distributed systems, hadoop, high performance computing, machine learning", pubstate = "published", tppubtype = "inproceedings", url = "https://md2k.org/images/papers/software/p1343-weimer_reef.pdf" }Matteo Interlandi, Kshitij Shah, Sai Deep Tetali, Muhammad Ali Gulzar, Seunghyun Yoo, Miryung Kim, Todd Millstein and Tyson Condie.
Titian: Data Provenance Support in Spark. Proc. VLDB Endow. 9(3):216–227, 2015. URL, DOI BibTeX@article{Interlandi:2015:TDP:2850583.2850595, author = "Interlandi, Matteo and Shah, Kshitij and Tetali, Sai Deep and Gulzar, Muhammad Ali and Yoo, Seunghyun and Kim, Miryung and Millstein, Todd and Condie, Tyson", title = "Titian: Data Provenance Support in Spark", journal = "Proc. VLDB Endow.", year = 2015, volume = 9, number = 3, pages = "216--227", issn = "2150-8097", abstract = "Debugging data processing logic in Data-Intensive Scalable Computing (DISC) systems is a difficult and time consuming effort. Today's DISC systems offer very little tooling for debugging programs, and as a result programmers spend countless hours collecting evidence (e.g., from log files) and performing trial and error debugging. To aid this effort, we built Titian, a library that enables data provenance---tracking data through transformations---in Apache Spark. Data scientists using the Titian Spark extension will be able to quickly identify the input data at the root cause of a potential bug or outlier result. Titian is built directly into the Spark platform and offers data provenance support at interactive speeds---orders-of-magnitude faster than alternative solutions---while minimally impacting Spark job performance; observed overheads for capturing data lineage rarely exceed 30% above the baseline job execution time.", doi = "https://doi.org/10.14778/2850583.2850595", publisher = "VLDB Endowment", pubstate = "published", tppubtype = "article", url = "https://md2k.org/images/papers/software/p216-interlandi_titian.pdf" }Salma Elmalaki, Lucas Wanner and Mani Srivastava.
CAreDroid: Adaptation Framework for Android Context-Aware Applications. In Proceedings of the 21st Annual International Conference on Mobile Computing and Networking. 2015, 386–399. URL, DOI BibTeX@inproceedings{Elmalaki:2015:CAF:2789168.2790108, author = "Elmalaki, Salma and Wanner, Lucas and Srivastava, Mani", title = "CAreDroid: Adaptation Framework for Android Context-Aware Applications", booktitle = "Proceedings of the 21st Annual International Conference on Mobile Computing and Networking", year = 2015, series = "MobiCom '15", pages = "386--399", address = "Paris, France", publisher = "ACM", abstract = "Context-awareness is the ability of software systems to sense and adapt to their physical environment. Many contemporary mobile applications adapt to changing locations, connectivity states, and available energy resources. Nevertheless, there is little systematic support for context-awareness in mobile operating systems. Because of this, application developers must build their own context-awareness adaptation engines, dealing directly with sensors and polluting application code with complex adaptation decisions. However, with adequate support from the runtime system, context monitoring could be performed efficiently in the background and adaptation could happen automatically [1]. Application developers would then only be required to implement methods tailored to different contexts. Just as file and socket abstractions help applications handle traditional input, output, and communication; a context-aware runtime system could help applications adapt according to user behavior and physical context.", doi = "10.1145/2789168.2790108", isbn = "978-1-4503-3619-2", keywords = "android, context-adaptation, context-aware computing", pubstate = "published", tppubtype = "inproceedings", url = "https://md2k.org/images/papers/software/p35-elmalaki_caredroid.pdf" }

